galaxy.hpc.ut.ee¶
Galaxy is an open source, web-based platform for data intensive biomedical research. It provides researchers with tools and computational resources through an easy-to-use interface.
Our Galaxy instance is tightly coupled to our main Rocket compute cluster. The jobs and workflows submitted through Galaxy will end up in the same queue as compute jobs submitted through our Rocket head node.
Request an account¶
Regardless of your affiliation, you can request access to Galaxy by filling out the form here .
Galaxy supports two ways to log in: using your University of Tartu credentials or through ETAIS (MyAccessID) authentication.
ETAIS¶
ETAIS users can log into Galaxy, but without explicitly asking for access, you will be in view only mode. While you can upload data, any jobs you try to run will fail.
As a prerequisite to using Galaxy, you should have a working Rocket (UT HPC) resource. This resource will be linked to your Galaxy account after requesting access. Rocket cluster resource ordering guide.
If you want to use Galaxy as an ETAIS user, then please send an email with your request to support@hpc.ut.ee . If you already have a Rocket resource, please include its name in your email.
University of Tartu¶
If you're already using the UT HPC cluster through the command line, you should be able to register automatically using your University of Tartu credentials.
For others, please use the form or send an email with your request to support@hpc.ut.ee . Make sure to include your UT account username.
Tools and datasets¶
Do keep in mind that UTHPC is in process of installing tools, sample datasets etc. You can send any requests regarding Galaxy choice of tools or reference datasets to support as well. You can view the tools available for installation into Galaxy at toolshed.g2.bx.psu.edu . Also, if you are planning to launch jobs on NGS datasets, please contact UTHPC first for support on tool profiling and resource allocation.
Importing data to Galaxy¶
There are multiple methods for uploading data to our Galaxy instance:
- Uploading through the web interface
- Importing from a URL
- Uploading via SSH/SFTP and importing through the web interface
Uploading through the web interface¶
This method is pretty straight forward and is good for uploading smaller files directly from your local storage.
First, locate and click the Upload Data button in the upper left corner:
This will open up the upload window:
Clicking Choose local file will open up your default file explorer where you can choose what files to upload. You can rename the file, choose what type of file it is and specify the genome. Start will begin the upload, the upload window can be closed after that. Upload progress and the uploaded data will appear in the active history.
Importing from a URL¶
If the data is readily available on a web server then Galaxy can directly download it from the URL. To import data from a URL, click the Paste/Fetch data in the upload window.
This will open up a text box where you can enter a URL or multiple URLs (one per line):
As before, Start will begin the import process.
The text box that Paste/Fetch data provides can also be used to directly paste the contents of a file to Galaxy, although this is not necessarily recommended, as having an actual copy of a file somewhere is better.
Uploading via SFTP¶
If the datasets are big or there are many of them then it is recommended to use SFTP to get them into Galaxy.
Prerequisites¶
For security and convenience it is recommended to use SSH key authentication to connect to Galaxy's SFTP server.
ETAIS users need to have a working FreeIPA account and SSH key configured in ETAIS self-service. The same procedure needs to be followed that gives access to the main Rocket cluster resources. FreeIPA configuration guide and ETAIS SSH key guide.
University users should first connect to our Rocket head node (rocket.hpc.ut.ee) using SSH and add the public key there. This process is roughly documented in this SSH access guide. If you can access rocket.hpc.ut.ee with an SSH key, then you should also be able to access Galaxy using the same key.
Using an FTP client¶
You can connect with Galaxy SFTP using any client that supports the protocol, the example is done using FileZilla.
Host: sftp://galaxy.hpc.ut.ee or sftp://m83.hpc.ut.ee, both names point to the same SFTP server.
Username: your University or ETAIS FreeIPA username, same username that is displayed in Galaxy
The password field can be left empty if you have SSH key authentication to rocket.hpc.ut.ee configured.
After connecting, a new folder is created, located in /gpfs/helios/galaxy/service/ftp/. The name of the folder is your Galaxy username. You can then use the SFTP client to upload your desired datasets.
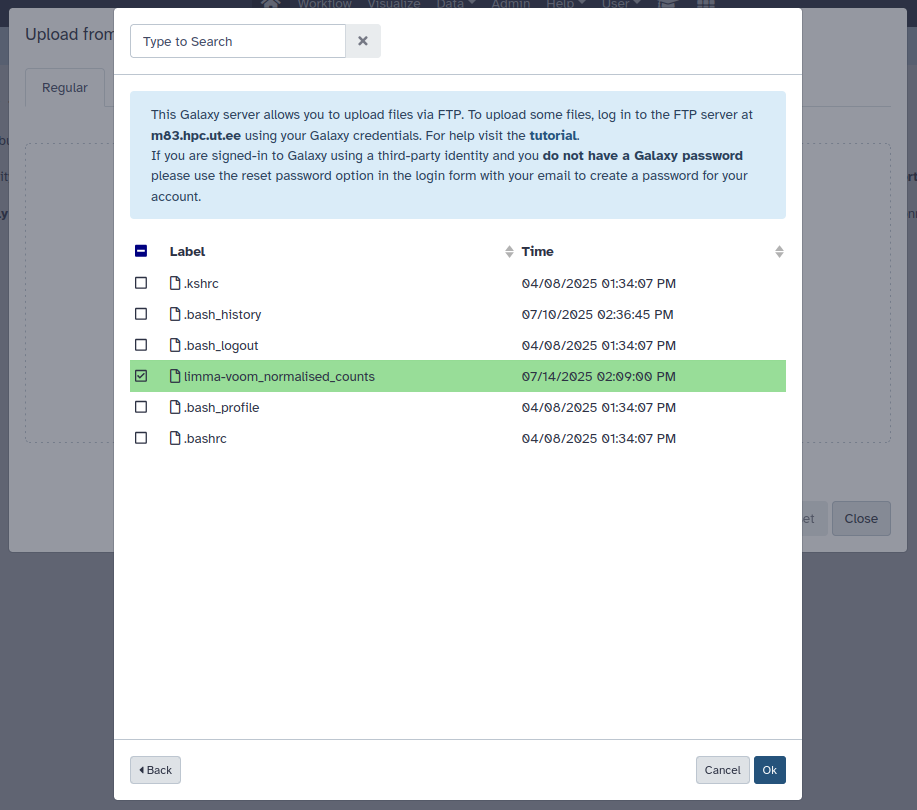
The datasets need to be imported from the web UI after the upload. For this, navigate back to the upload window in Galaxy and press Choose remote files. This will open another window:
From here click FTP Directory. You will be presented the contents of your folder, choose the datasets you want to import and press Ok:
As before, Start will begin the import process.
Other methods¶
If you are comfortable with the command line interface, then there is a plethora of other file transfer tools that can be used over SSH. For inspiration, you can check out our general file transfer guide. Just keep in mind, that the path on Galaxy will be /gpfs/helios/galaxy/service/ftp/<your_username> and the files need to be imported into Galaxy through the web interface.